Tools and Databases
-
CCharPPI
Computational Characterisation of Protein-Protein Interactions.
-
OPRA Server
OPRA (Optimal Protein-RNA Area) is useful for identifying potential RNA-binding sites on proteins and can help to model protein-RNA interactions of biological and therapeutic interest.
-
Protein-RNA Benchmark
The Protein-RNA benchmark v1.0 is composed of 106 cases, with 5 unbound-unbound, 4 unbound-pseudo-unbound, 62 unbound-bound, 5 unbound-model, 8 model-unbound, 19 model-bound cases and 3 model-model.
-
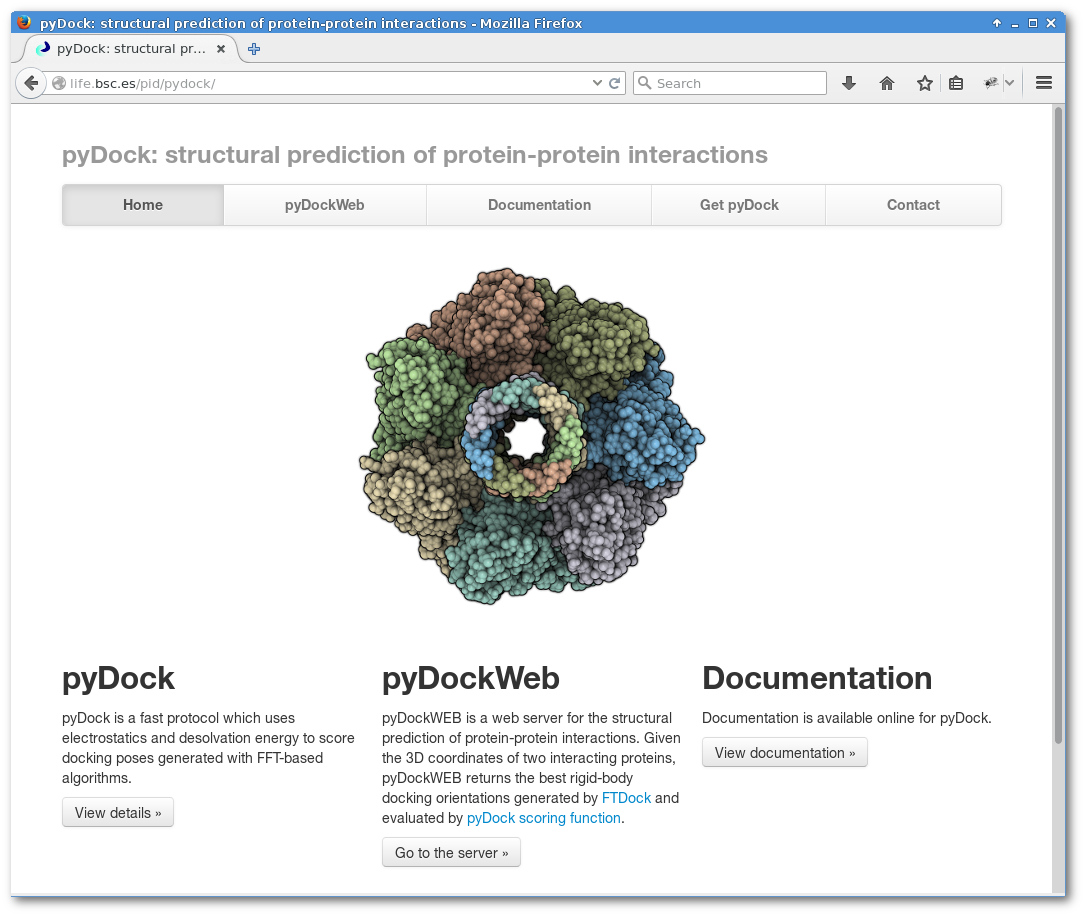
pyDock
pyDock is a fast protocol which uses electrostatics and desolvation energy to score docking poses generated with FFT-based algorithms.
-
pyDockEneRes
pyDock Energy per Residue
-
pyDockRescoring
PyDockRescoring is a web server for rescoring of pyDockWEB server jobs.
-
pyDockSAXS
pyDockSAXS server is a web server for rigid-body protein-protein docking that combines computational and experimental information.
-
pyDockWEB
pyDockWEB is a web server for the structural prediction of protein-protein interactions.
-
SKEMPI2
The new version of the SKEMPI database, with more than 7000 mutation data. The SKEMPI2 database contains data on the changes in thermodynamic parameters and kinetic rate constants upon mutation, for protein-protein interactions for which a structure of the complex has been solved and is available in the protein databank.
Server status
- CCharPPI - online
- OPRA - online
- pyDockEneRes - online
- pyDockRescoring - online
- pyDockSAXS - online
- pyDockWEB - online
Queue status
- pyDockWEB
Running(3) / Queued(1)